Publications
Recent research and discoveries from our team.
2025
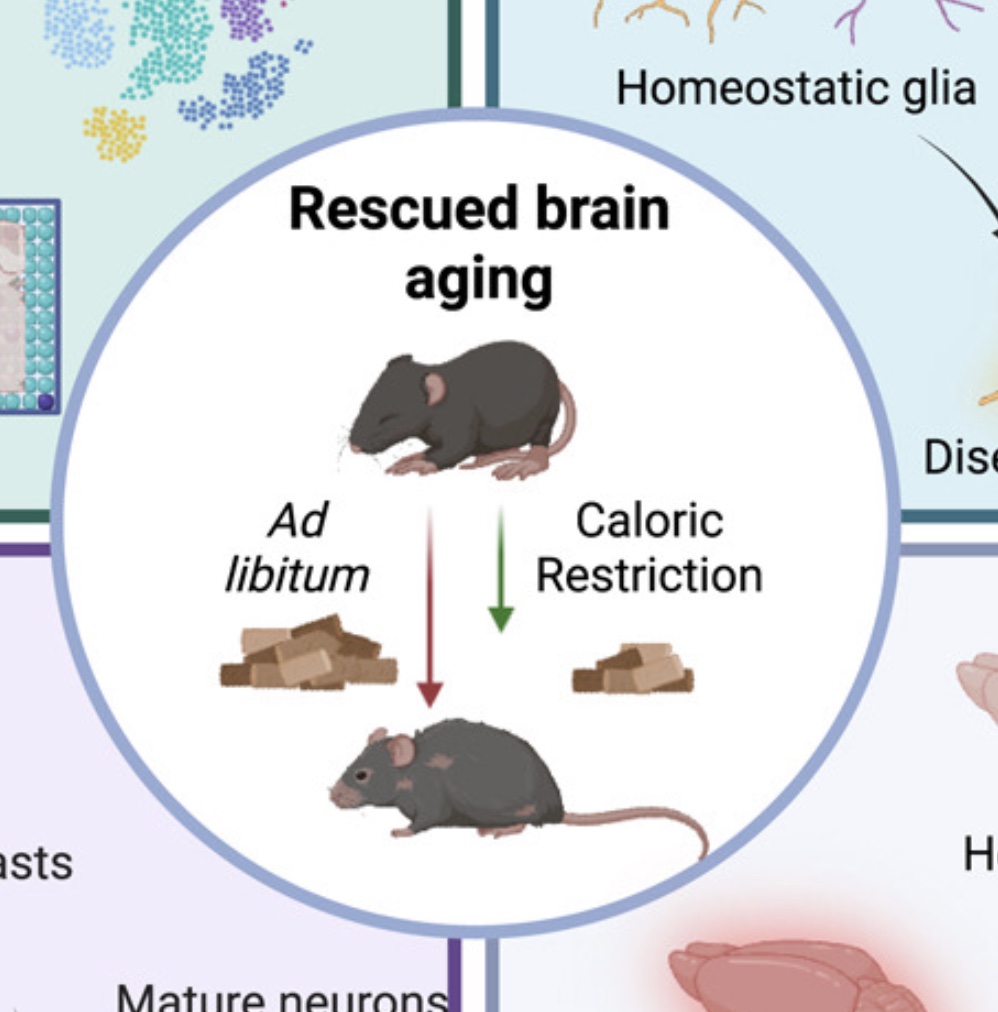
Spatiotemporal profiling reveals the impact of caloric restriction in the aging mammalian brain
Zhang Z, Epstein A, Schaefer C, Abdulraouf A, Jiang W, Zhou W, Cao J
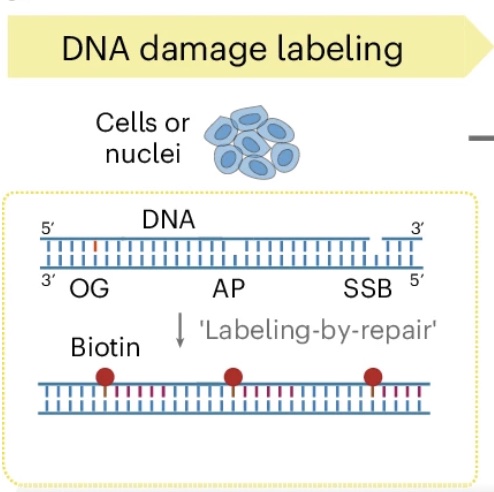
Single-cell parallel analysis of DNA damage and transcriptome reveals selective genome vulnerability
Bai D, Cao Z, Attada N, Song J, Zhu C
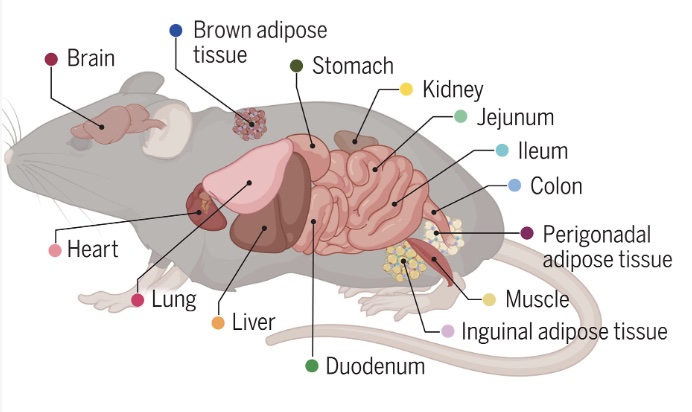
A panoramic view of cell population dynamics in mammalian aging
Zhang Z, Schaefer C, Jiang W, Lu Z, Lee J, Sziraki A, Abdulraouf A, Wick B, Haeussler M, Li Z, Molla G, Satija R, Zhou W, Cao J

Large-scale single-cell phylogenetic mapping of clonal evolution in the human aging esophagus
Prieto T, Yuan DJ, Zinno J, Hughes C, Midler N, Kao S, Huuhtanen J, Raviram R, Fotopoulou F, Ruthen N, Rajagopalan S, Schiffman JS, D'Avino AR, Yoon S-H, Sotelo J, Omans ND, Wheeler N, Garces A, Pradhan B, Cheng AP, Robine N, Potenski C, Godfrey K, Kakiuchi N, Yokoyama A, Ogawa S, Abrams J, Raimondi I, Landau DA
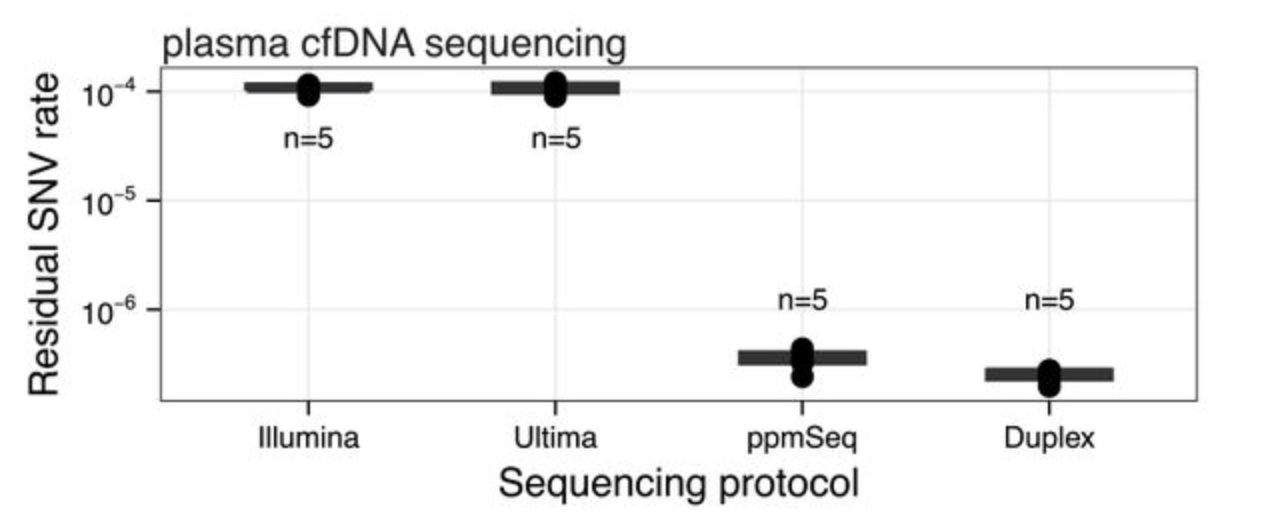
Paired plus-minus sequencing is an ultra-high throughput and accurate method for dual strand sequencing of DNA molecules
Cheng AP, Rusinek I, Sossin A, Widman AJ, Meiri E, Krieger G, Hirschberg O, Shem Tov D, Gilad S, Jaimovich A, Barad O, Avaylon S, Rajagopalan S, Potenski C, Prieto T, Yuan DJ, Furatero R, Runnels A, Costa BM, Shoag JE, Al Assaad M, Sigouros M, Manohar J, King A, Wilkes D, Otilano J, Malbari MS, Elemento O, Mosquera JM, Altorki NK, Saxena A, Callahan MK, Robine N, Germer S, Evrony GD, Faltas BM, Landau DA

Mapping transcriptional responses to cellular perturbation dictionaries with RNA fingerprinting
Grabski IN, Lee J, Blair JD, Dalgarno C, Mascio I, Bradu A, Knowles DA, Satija R

Iterative, multimodal, and scalable single-cell profiling for discovery and characterization of signaling regulators
Blair JD, Bradu A, Dalgarno C, Grabski IN, Satija R
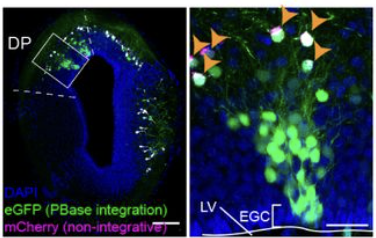
A conserved logic for the development of cortical layering in tetrapods
Deryckere A, Choudhary S, Lynch C, Limperis L, Affatato P, Woych J, Gumnit E, Gurrola A, Satija R, Mayer C, Tosches M
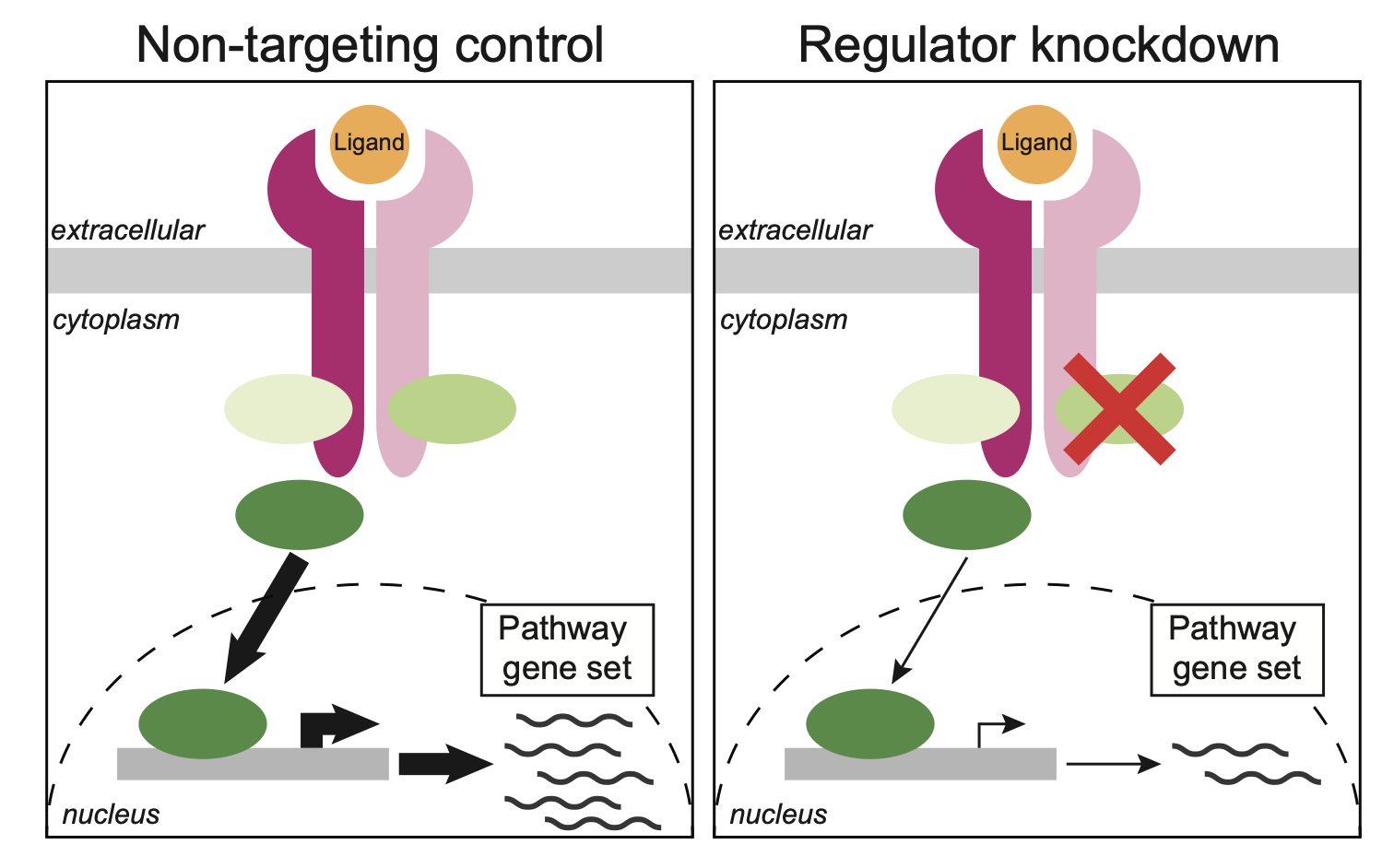
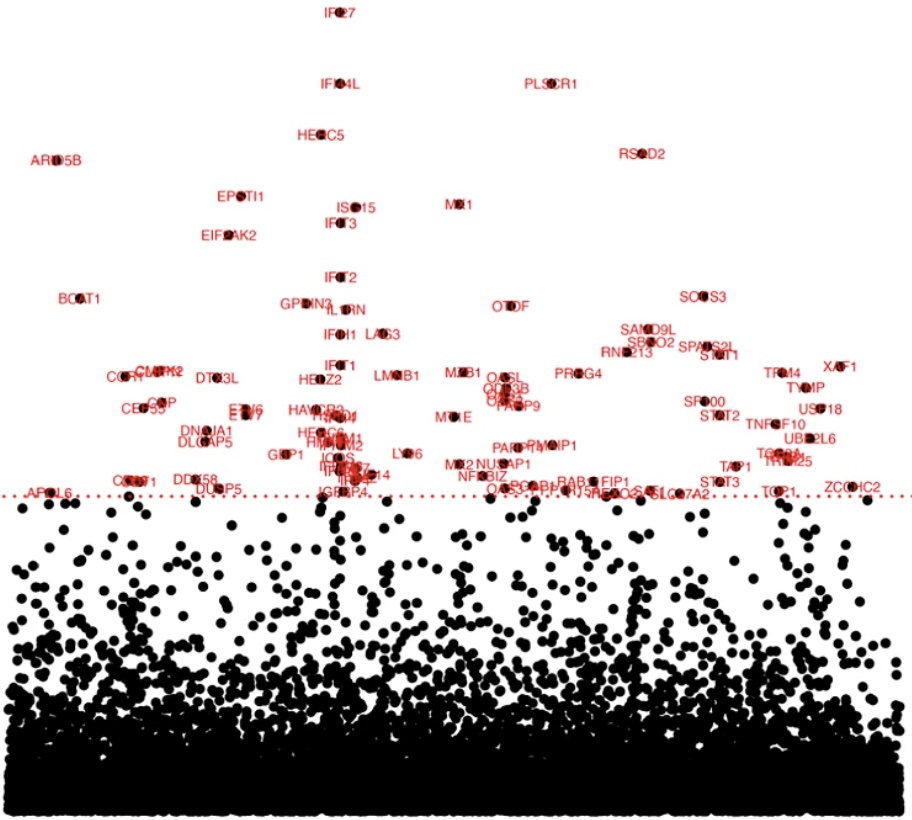
Systematic reconstruction of molecular pathway signatures using scalable single-cell perturbation screens
Jiang L*, Dalgarno C*, Papalexi E, Mascio I, Wessels HH, Yun H, Iremadze N, Lithwick-Yanai G, Lipson D, Satija R
Improving reproducibility of differentially expressed genes in single-cell transcriptomic studies of neurodegenerative diseases through meta-analysis
Nakatsuka N, Adler D, Jiang L, Hartman A, Cheng E, Klann E, Satija R
2024
Defining heritability, plasticity, and transition dynamics of cellular phenotypes in somatic evolution
Schiffman JS, D'Avino AR, Prieto T, Pang Y, Fan Y, Rajagopalan S, Potenski C, Hara T, Suvà ML, Gawad C, Landau DA
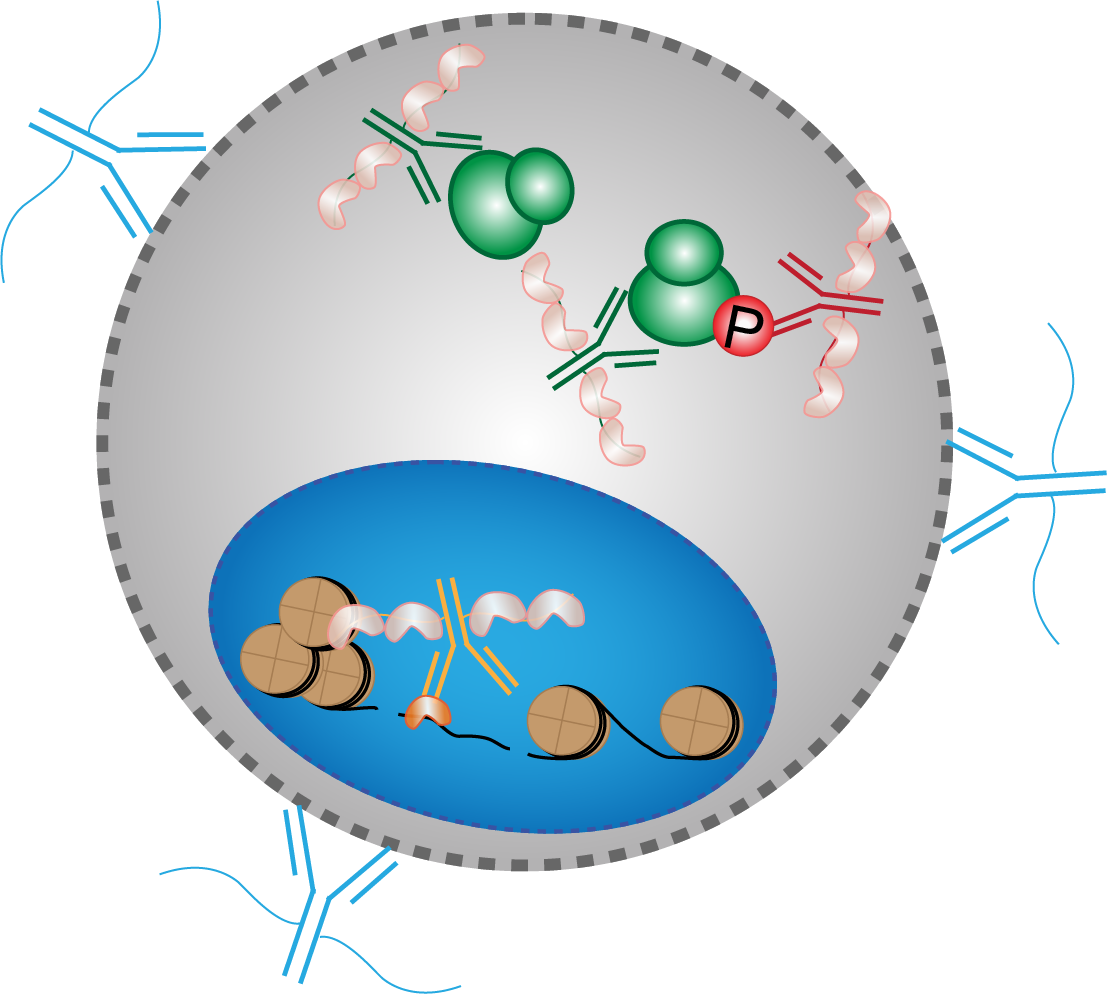
Phospho-seq: Integrated, multi-modal profiling of intracellular protein dynamics in single cells
John D. Blair, Austin Hartman, Fides Zenk, Carol Dalgarno, Barbara Treutlein, Rahul Satija
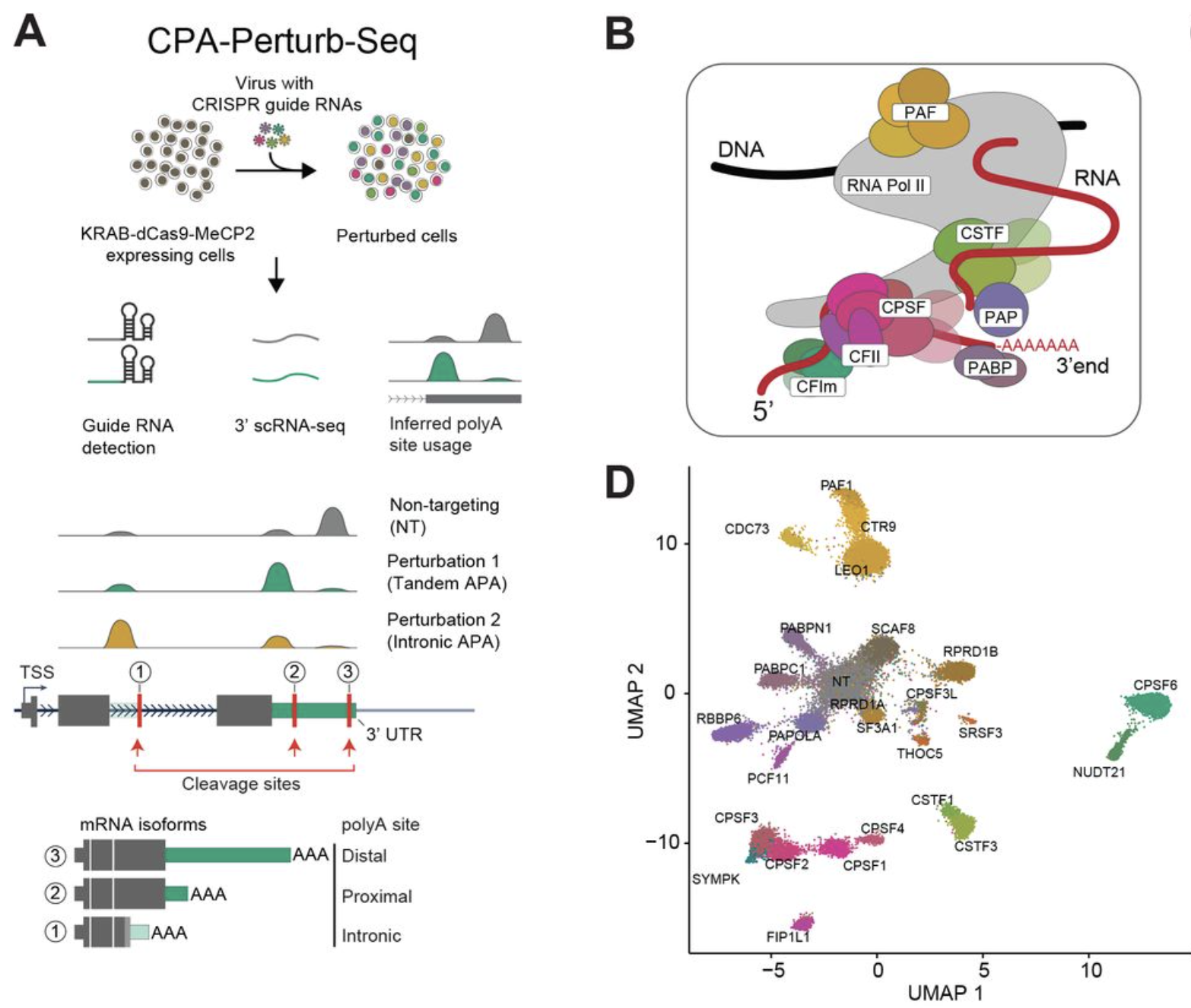
CPA-Perturb-seq: Multiplexed single-cell characterization of alternative polyadenylation regulators
Madeline H. Kowalski, Hans-Hermann Wessels, Johannes Linder, Saket Choudhary, Austin Hartman, Yuhan Hao, Isabella Mascio, Carol Dalgarno, Anshul Kundaje, Rahul Satija
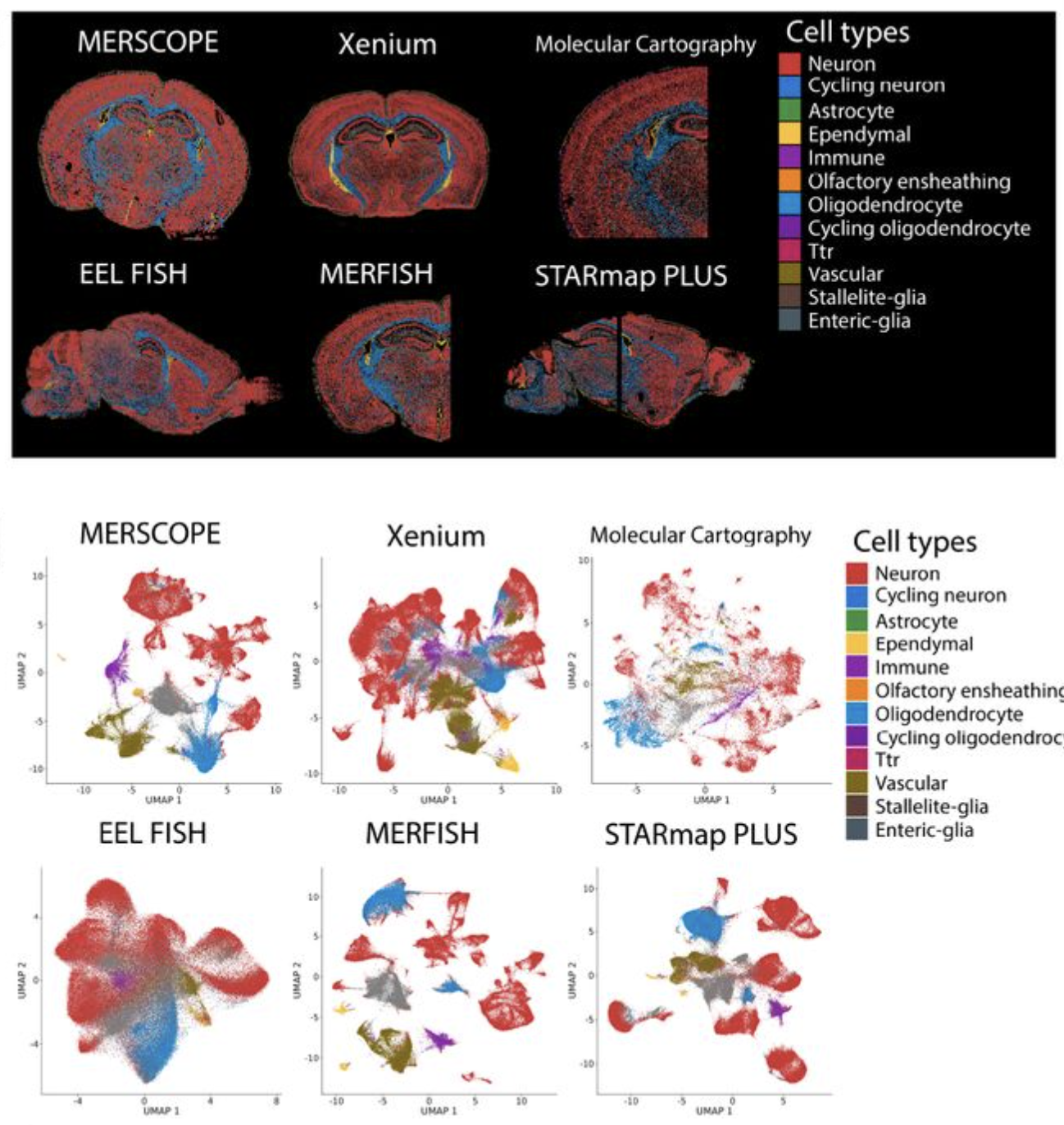
Comparative analysis of multiplexed in situ gene expression profiling technologies
Austin Hartman, Rahul Satija
2023
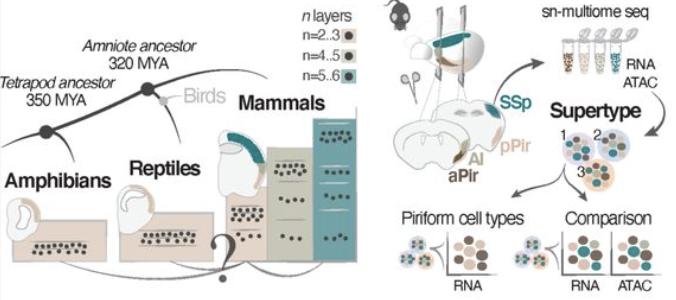
Mammalian olfactory cortex neurons retain molecular signatures of ancestral cell types
S. Zeppilli, A. Ortega Gurrola, P. Demetci, D. H. Brann, R. Attey, N. Zilkha, T. Kimchi, S. R. Datta, R. Singh, M. A. Tosches, A. Crombach, A. Fleischmann
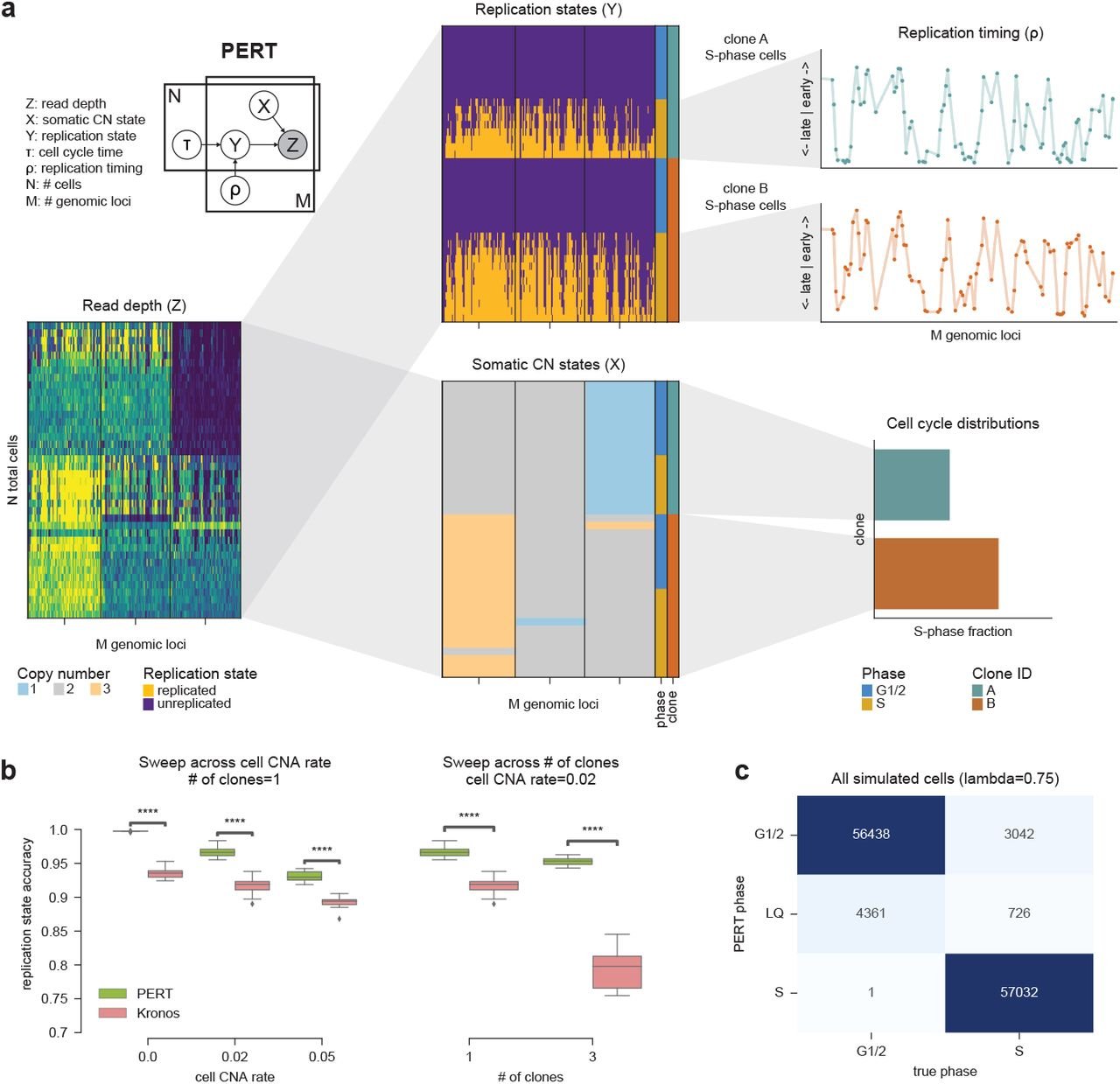
Single-cell DNA replication dynamics in genomically unstable cancers
Adam C. Weiner, Marc J. Williams, Hongyu Shi, Ignacio Vázquez-García, Sohrab Salehi, Nicole Rusk, Samuel Aparicio, Sohrab P. Shah, Andrew McPherson
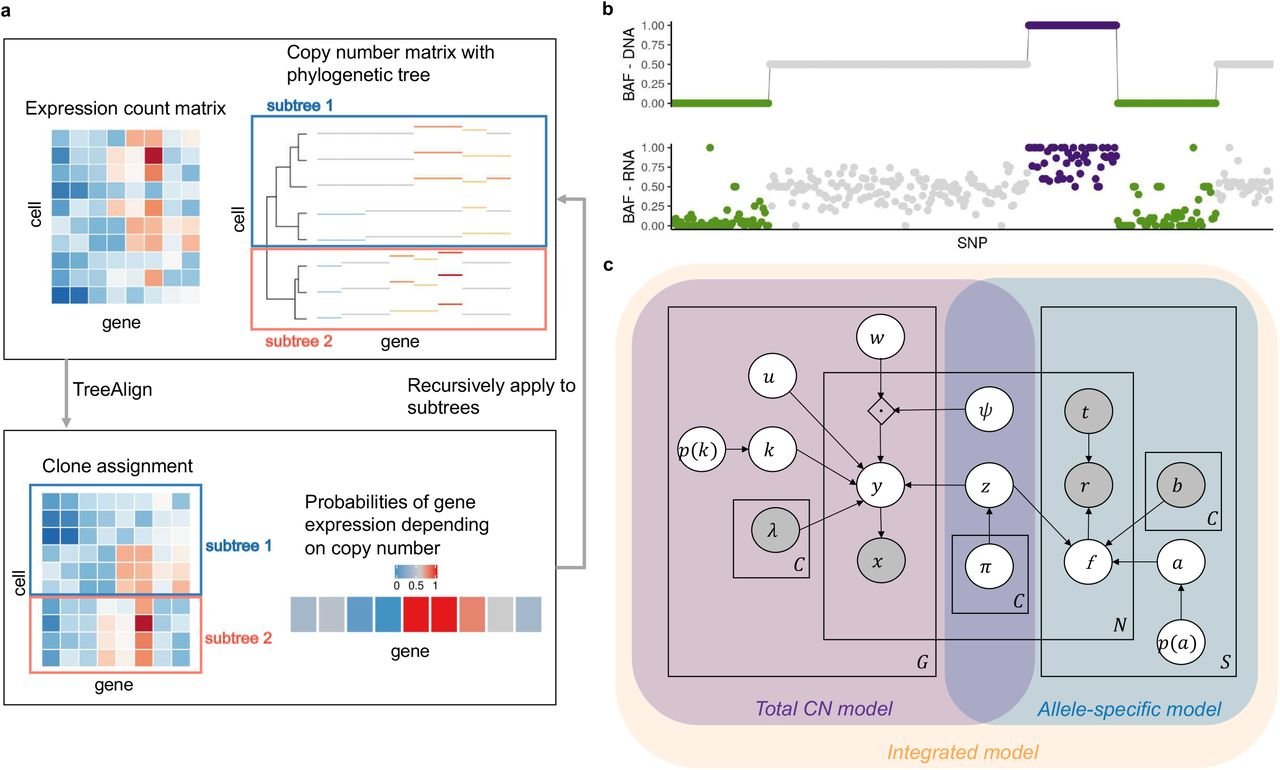
Exploiting allele-specific transcriptional effects of subclonal copy number alterations for genotype-phenotype mapping in cancer cell populations
Hongyu Shi, Marc J. Williams, Gryte Satas, Adam C. Weiner, Andrew McPherson, Sohrab P. Shah
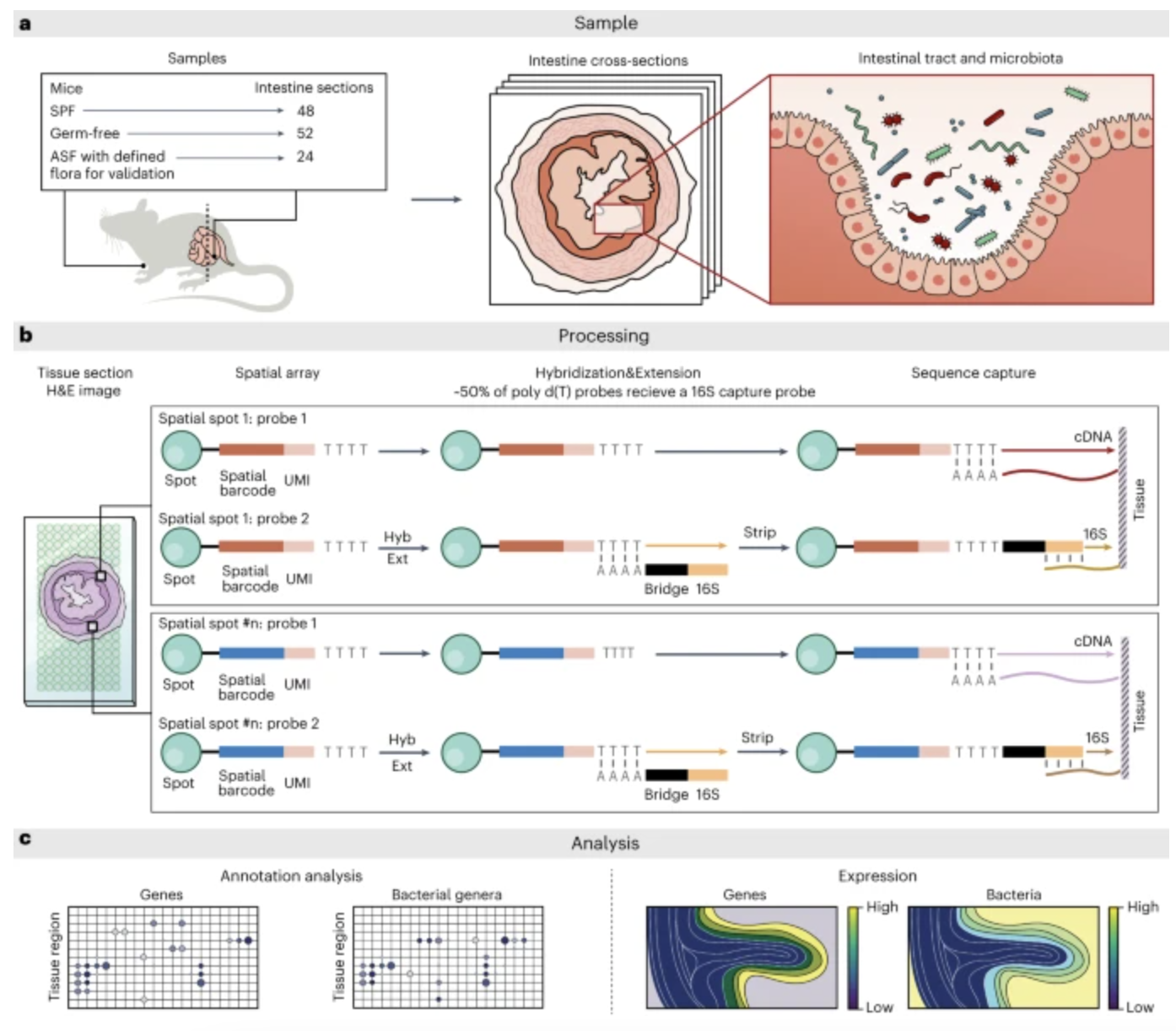
Spatial host-microbiome sequencing reveals niches in the mouse gut
Britta Lötstedt, Martin Stražar, Ramnik Xavier, Aviv Regev & Sanja Vickovic
A global view of aging and Alzheimer’s pathogenesis-associated cell population dynamics and molecular signatures in human and mouse brains
Andras Sziraki, Ziyu Lu, Jasper Lee, Gabor Banyai, Sonya Anderson, Abdulraouf Abdulraouf, Eli Metzner, Andrew Liao, Jason Banfelder, Alexander Epstein, Chloe Schaefer, Zihan Xu, Zehao Zhang, Li Gan, Peter T. Nelson, Wei Zhou & Junyue Cao
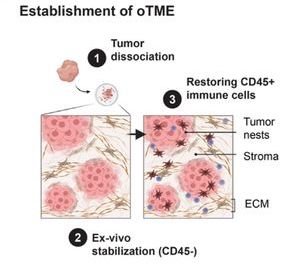
Breast Cancer Macrophage Heterogeneity and Self-renewal are Determined by Spatial Localization
Nir Ben-Chetrit, Xiang Niu, Jesus Sotelo, Ariel D. Swett, Vinagolu K. Rajasekhar, Maria S. Jiao, Caitlin M. Stewart, Priya Bhardwaj, Sanjay Kottapalli, Saravanan Ganesan, Pierre-Louis Loyher, Catherine Potenski, Assaf Hannuna, Kristy A. Brown, Neil M. Iyengar, Dilip D. Giri, Scott W. Lowe, John H. Healey, Frederic Geissmann, Irit Sagi, Johanna A. Joyce, Dan A. Landau
Tracking cell-type-specific temporal dynamics in human and mouse brains
Ziyu Lu, Melissa Zhang, Jasper Lee, Andras Sziraki, Sonya Anderson, Zehao Zhang, Zihan Xu, Weirong Jiang, Shaoyu Ge, Peter T. Nelson, Wei Zhou, Junyue Cao
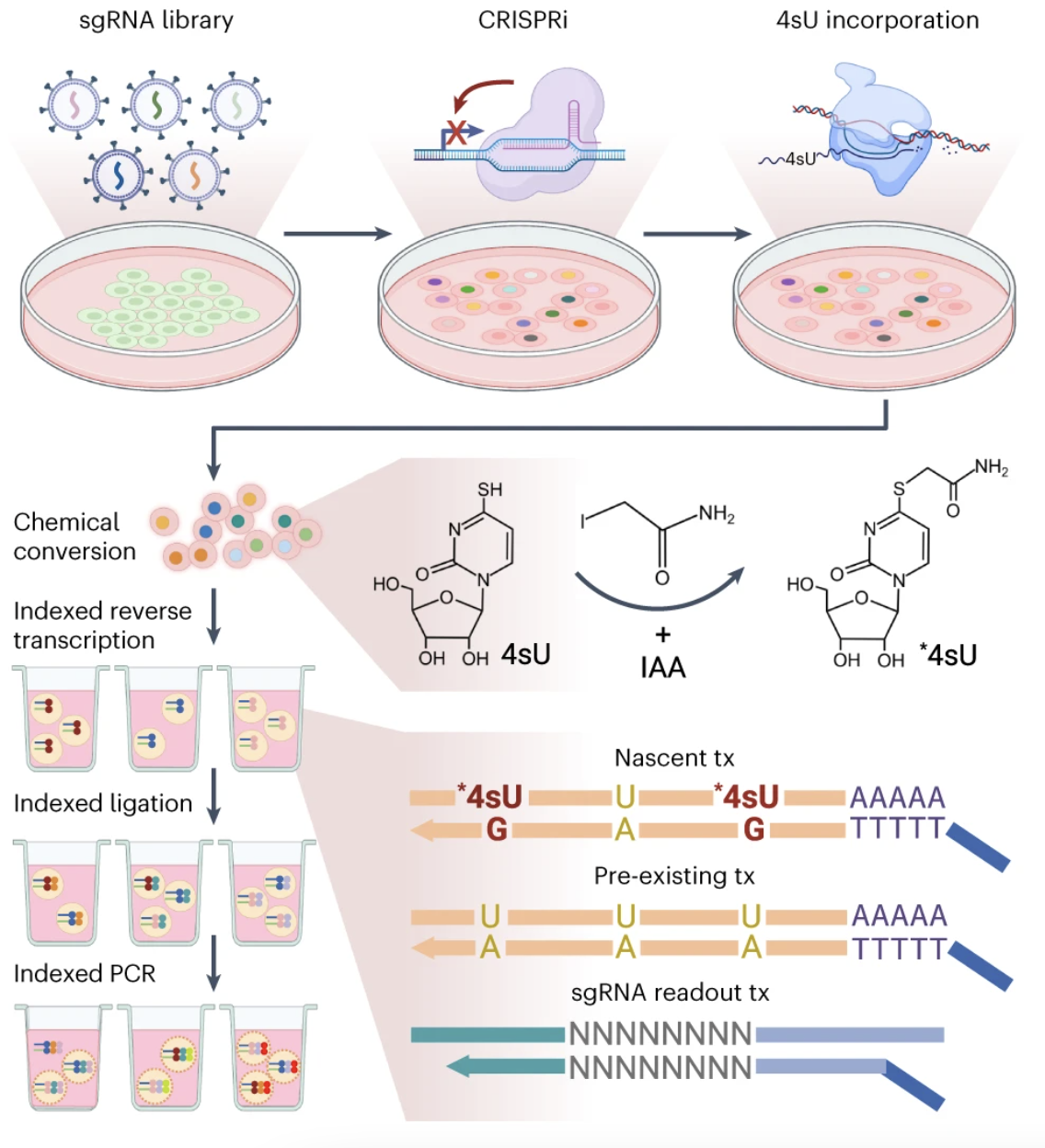
Dissecting key regulators of transcriptome kinetics through scalable single-cell RNA profiling of pooled CRISPR screens
Zihan Xu, Andras Sziraki, Jasper Lee, Wei Zhou, Junyue Cao
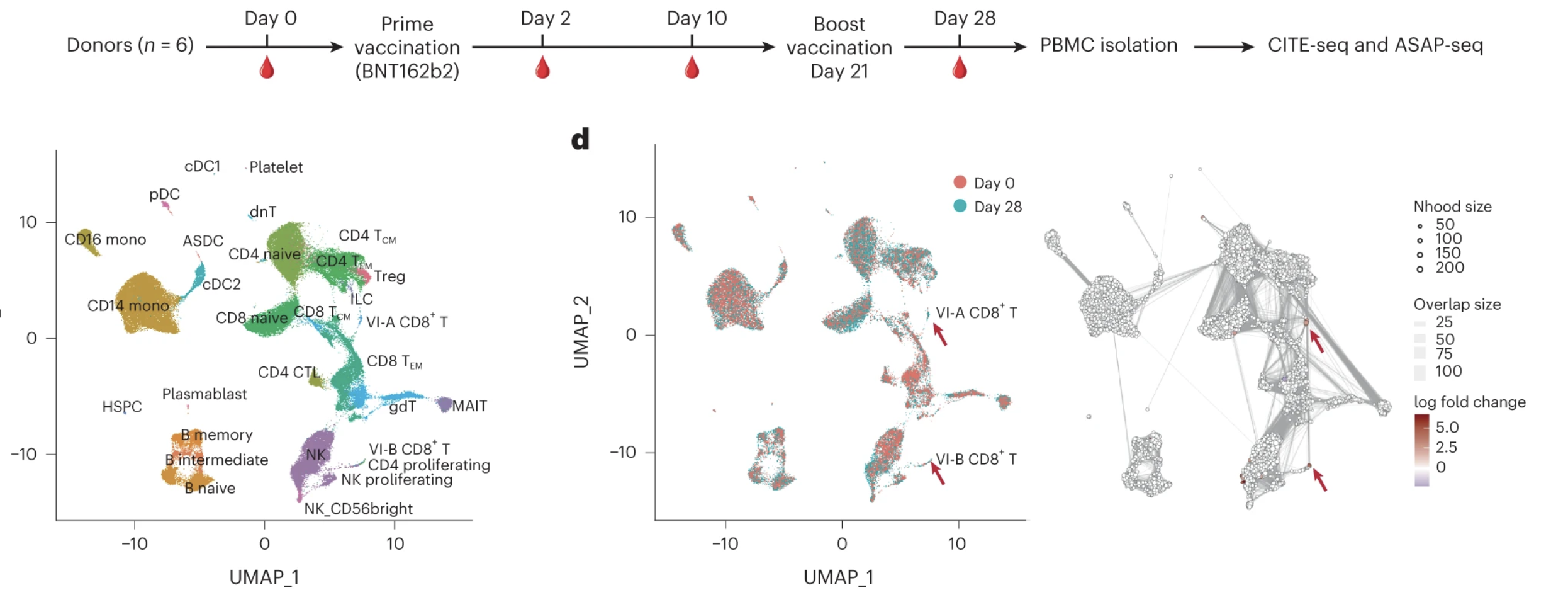
Multimodal single-cell datasets characterize antigen-specific CD8+ T cells across SARS-CoV-2 vaccination and infection
Bingjie Zhang, Rabi Upadhyay, Yuhan Hao, Marie I. Samanovic, Ramin S. Herati, John D. Blair, Jordan Axelrad, Mark J. Mulligan, Dan R. Littman & Rahul Satija
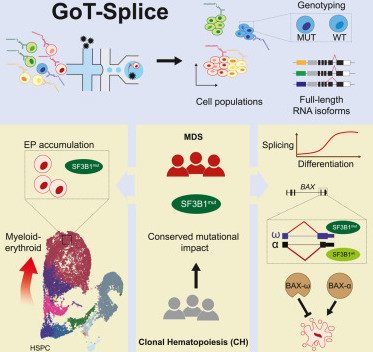
Single-cell multi-omics defines the cell-type-specific impact of splicing aberrations in human hematopoietic clonal outgrowths
Mariela Cortés-López, Paulina Chamely, Allegra G. Hawkins, Robert F. Stanley, Ariel D. Swett, Saravanan Ganesan, Tarek H. Mouhieddine, Xiaoguang Dai, Lloyd Kluegel, Celine Chen, Kiran Batta, Nili Furer, Rahul S. Vedula, John Beaulaurier , Alexander W. Drong, Scott Hickey , Neville Dusaj, Gavriel Mullokandov, Adam M. Stasiw, Jiayu Su, Dan A. Landau
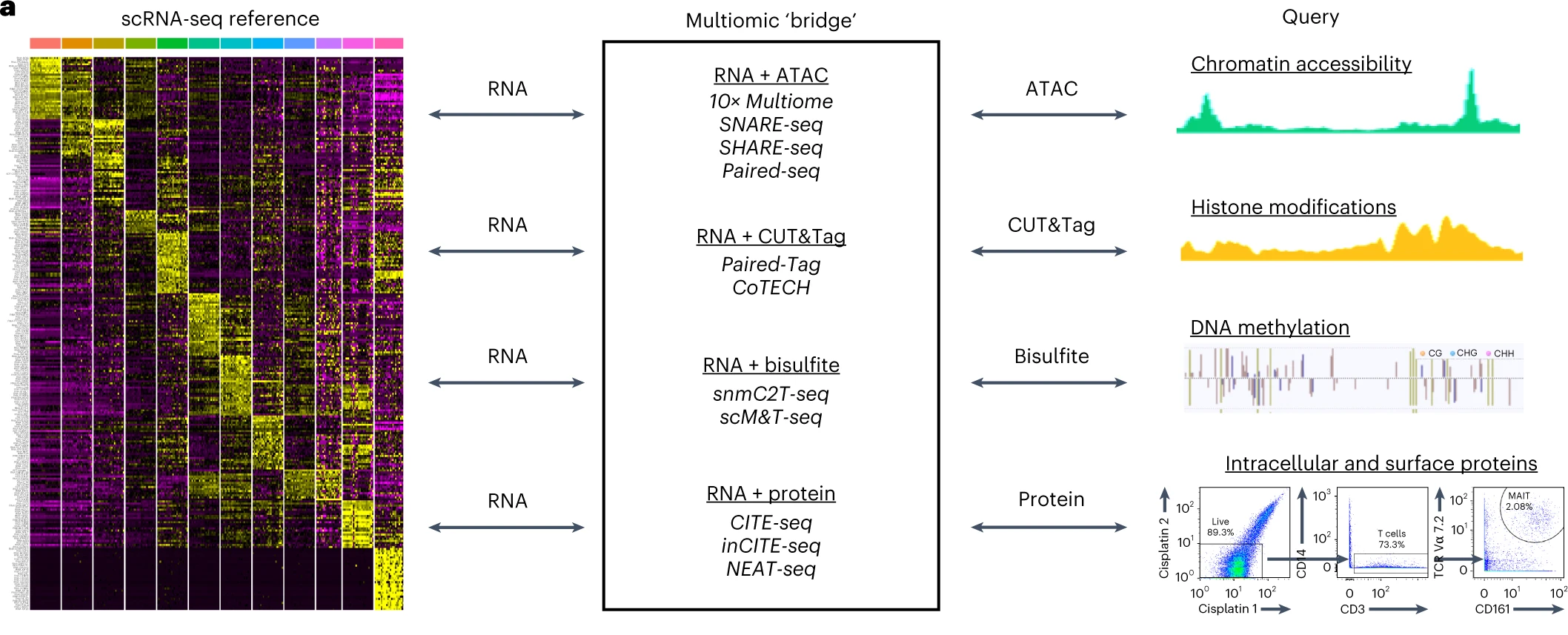
Dictionary learning for integrative, multimodal, and scalable single-cell analysis
Yuhan Hao, Tim Stuart, Madeline Kowalski, Saket Choudhary, Paul Hoffman, Austin Hartman, Avi Srivastava, Gesmira Molla, Shaista Madad, Carlos Fernandez-Granda, Rahul Satija

Defining ancestry, heritability and plasticity of cellular phenotypes in somatic evolution
Joshua S. Schiffman, Andrew R. D’Avino, Tamara Prieto, Catherine Potenski, Yilin Fan, Toshiro Hara, Mario L. Suvà, Dan A. Landau
2022

Efficient combinatorial targeting of RNA transcripts in single cells with Cas13 RNA Perturb-seq
Hans-Hermann Wessels, Alejandro Méndez-Mancilla, Yuhan Hao, Efthymia Papalexi, William M. Mauck III, Lu Lu, John A. Morris, Eleni P. Mimitou, Peter Smibert, Neville E. Sanjana & Rahul Satija
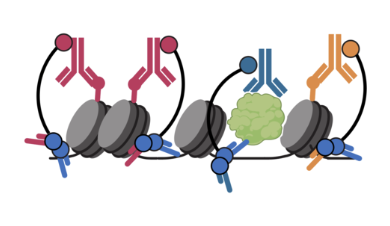
Nanobody-tethered transposition enables multifactorial chromatin profiling at single-cell resolution
Tim Stuart, Stephanie Hao, Bingjie Zhang, Levan Mekerishvili, Dan A. Landau, Silas Maniatis, Rahul Satija & Ivan Raimondi
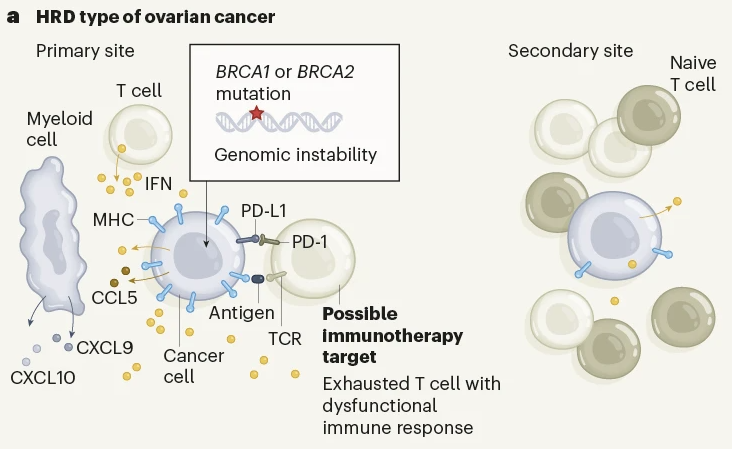
Ovarian cancer mutational processes drive site-specific immune evasion
Ignacio Vázquez-García, Florian Uhlitz, Nicholas Ceglia, Jamie L. P. Lim, Michelle Wu, Neeman Mohibullah, Juliana Niyazov, Arvin Eric B. Ruiz, Kevin M. Boehm, Viktoria Bojilova, Christopher J. Fong, Tyler Funnell, Diljot Grewal, Eliyahu Havasov, Samantha Leung, Arfath Pasha, Druv M. Patel, Maryam Pourmaleki, Nicole Rusk, Hongyu Shi, Rami Vanguri, Marc J. Williams, Allen W. Zhang, Vance Broach, …Sohrab P. Shah
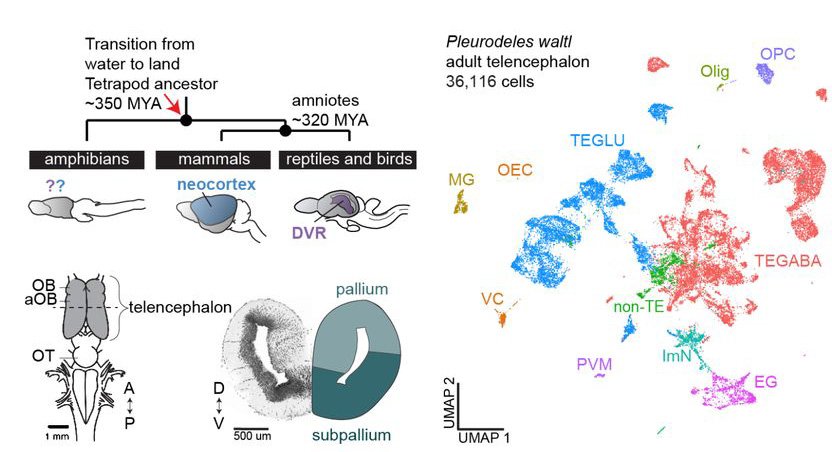
Cell type profiling in salamanders identifies innovations in vertebrate forebrain evolution
Jamie Woych, Alonso Ortega Gurrola, Astrid Deryckere, Eliza C. B. Jaeger, Elias Gumnit, Gianluca Merello, Jiacheng Gu, Alberto Joven Araus, Nicholas D. Leigh, Maximina Yun, András Simon, Maria Antonietta Tosches
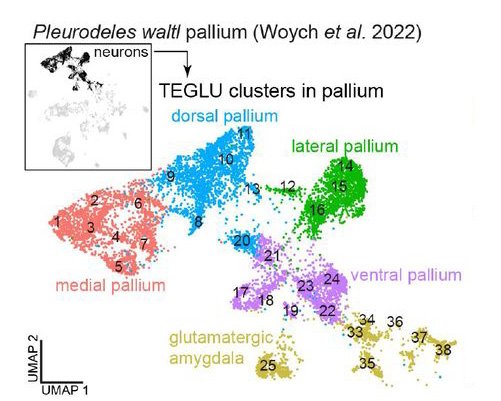
Glutamatergic neuron types in the amygdala of the urodele amphibian Pleurodeles waltl
Astrid Deryckere, Jamie Woych, Eliza C. B. Jaeger, Maria Antonietta Tosches
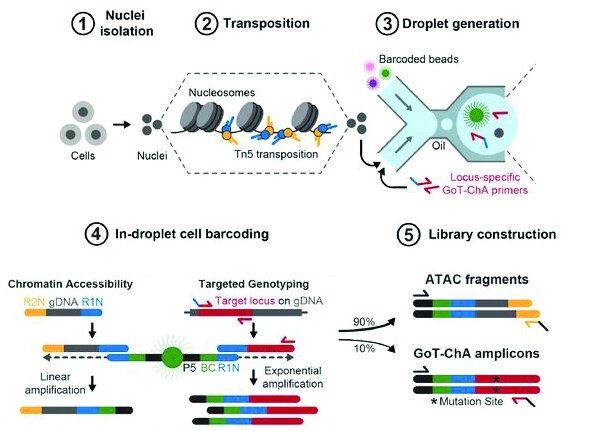
Integrated Single-Cell Genotyping and Chromatin Accessibility Charts JAK2V617F Human Hematopoietic Differentiation
Robert M. Myers, Franco Izzo, Sanjay Kottapalli, Tamara Prieto, Andrew Dunbar, Robert L. Bowman, Eleni P. Mimitou, Maximilian Stahl, Sebastian El Ghaity-Beckley, JoAnn Arandela, Ramya Raviram, Saravanan Ganesan, Levan Mekerishvili, Ronald Hoffman, Ronan Chaligné, Omar Abdel-Wahab, Peter Smibert, Bridget Marcellino, Ross L. Levine, Dan Avi Landau

Integrated protein and transcriptome high-throughput spatial profiling
Nir Ben Chetrit, Xiang Niu, Ariel D. Swett, Jesus Sotelo, Maria S. Jiao, Patrick Roelli, Marlon Stoeckius, Dan Avi Landau
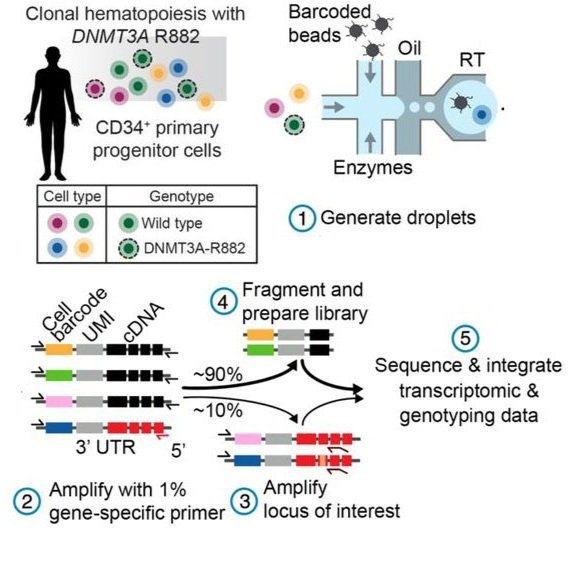
Single-cell multi-omics of human clonal hematopoiesis reveals that DNMT3A R882 mutations perturb early progenitor states through selective hypomethylation
Anna S. Nam, Neville Dusaj, Franco Izzo, Rekha Murali, Robert M. Myers, Tarek Mouhieddine, Jesus Sotelo, Salima Benbarche, Michael Waarts, Federico Gaiti, Sabrin Tahri, Ross Levine, Omar Abdel-Wahab, Lucy A. Godley, Ronan Chaligne, Irene Ghobrial, Dan A. Landau
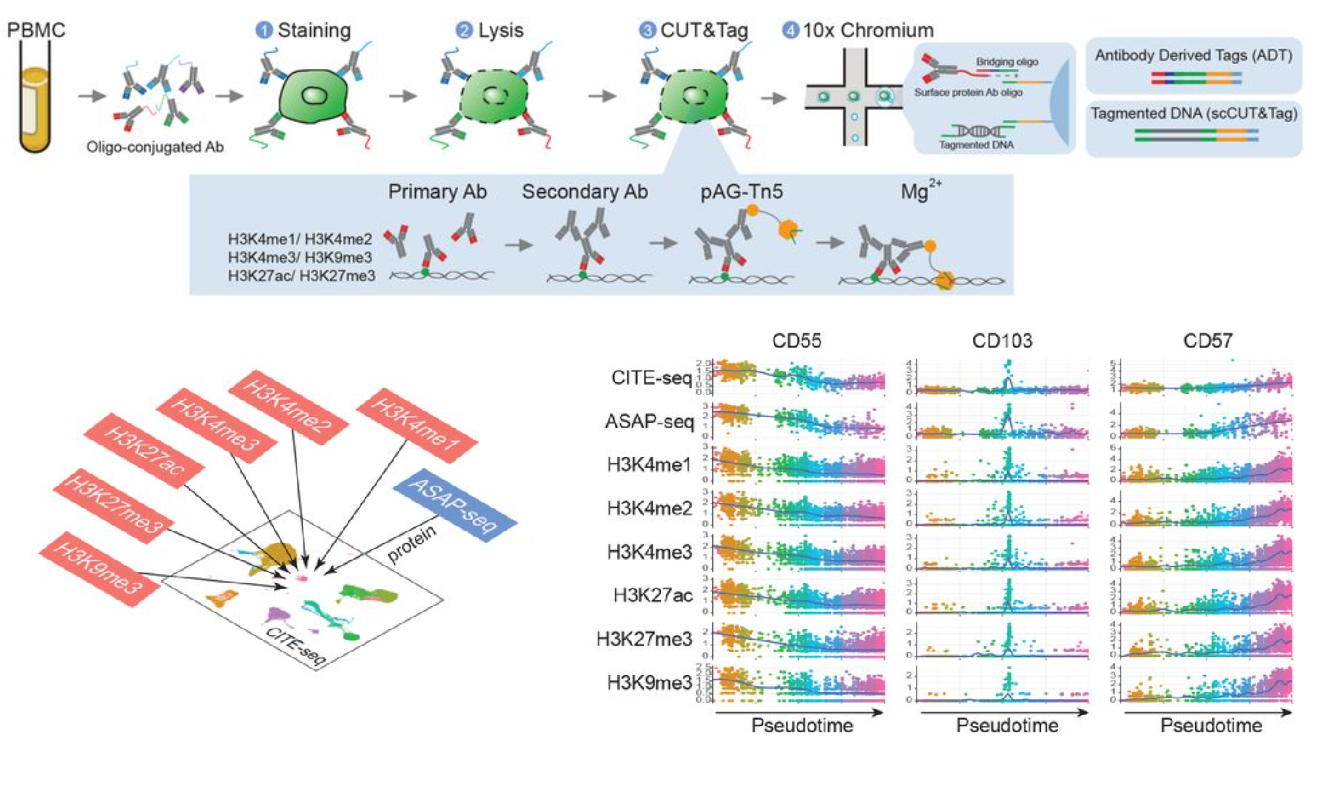
Characterizing cellular heterogeneity in chromatin state with scCUT&Tag-pro
Bingjie Zhang, Avi Srivastava, Eleni Mimitou, Tim Stuart, Ivan Raimondi, Yuhan Hao, Peter Smibert, Rahul Satija
PySeq2500: An open source toolkit for repurposing HiSeq 2500 sequencing systems as versatile fluidics and imaging platforms
Kunal Pandit, Joana Petrescu, Miguel Cuevas, William Stephenson, Peter Smibert, Hemali Phatnani, Silas Maniatis
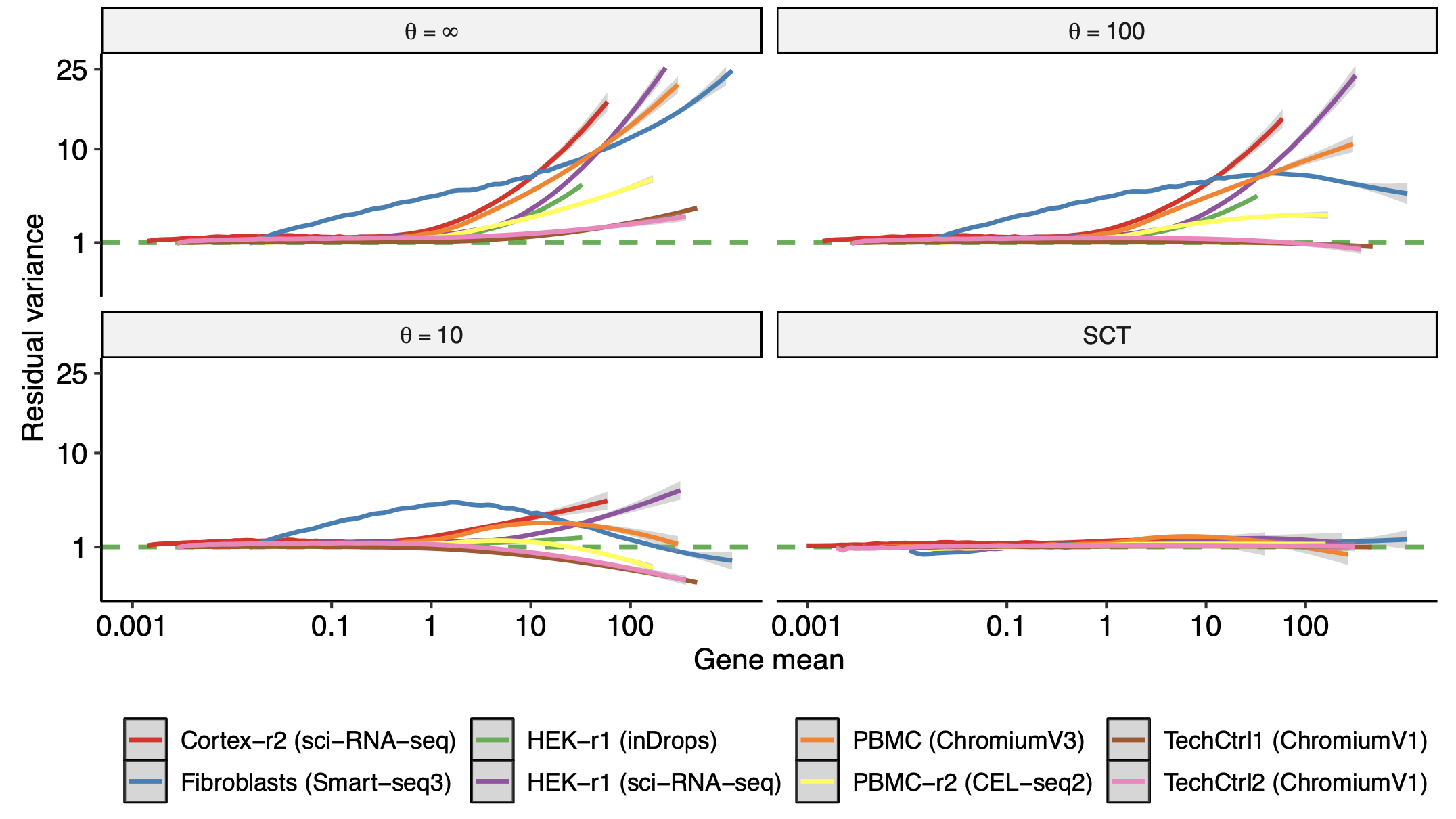
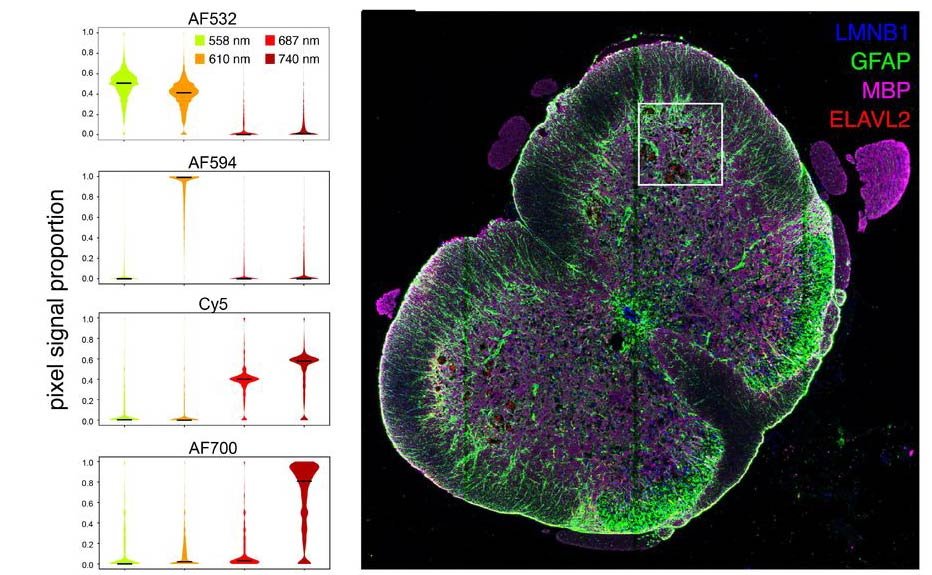
Comparison and evaluation of statistical error models for scRNA-seq
Saket Choudhary, Rahul Satija
2021

Multimodal single-cell chromatin analysis with Signac
Tim Stuart, Avi Srivastava, Shaista Madad, Caleb Lareau, Rahul Satija
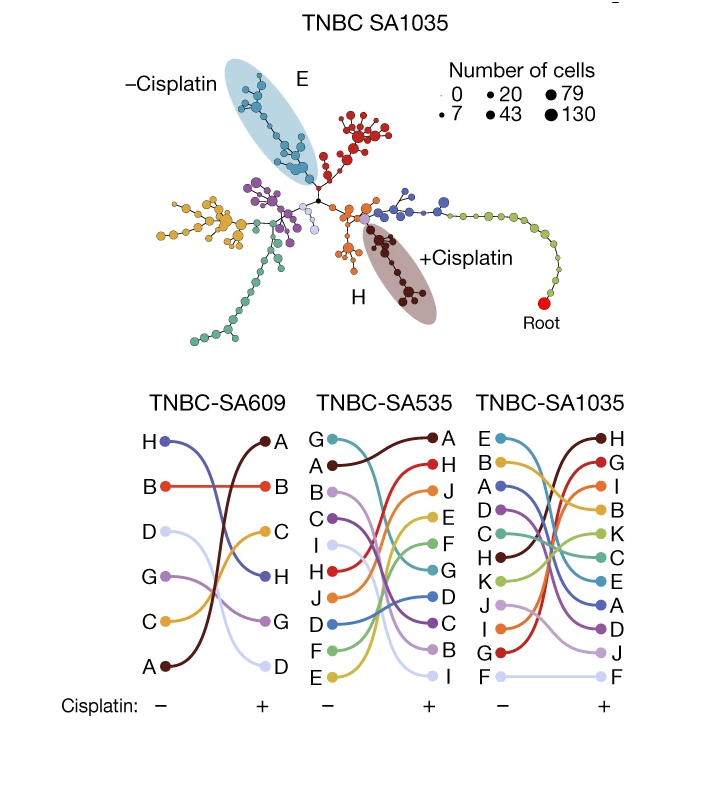
Clonal fitness inferred from time-series modelling of single-cell cancer genomes
Sohrab Salehi, Farhia Kabeer, Nicholas Ceglia, Mirela Andronescu, Marc J. Williams, Kieran R. Campbell, Tehmina Masud, Beixi Wang, Justina Biele, Jazmine Brimhall, David Gee, Hakwoo Lee, Jerome Ting, Allen W. Zhang, Hoa Tran, Ciara O’Flanagan, Fatemeh Dorri, Nicole Rusk, Teresa Ruiz de Algara, So Ra Lee, Brian Yu Chieh Cheng, Peter Eirew, Takako Kono, Jenifer Pham, Diljot Grewal, Daniel Lai, Richard Moore, Andrew J. Mungall, Marco A. Marra, IMAXT Consortium, Andrew McPherson, Alexandre Bouchard-Côté, Samuel Aparicio & Sohrab P. Shah

A convolutional neural network for common coordinate registration of high-resolution histology images
Aidan C Daly, Krzysztof J Geras, Richard A Bonneau
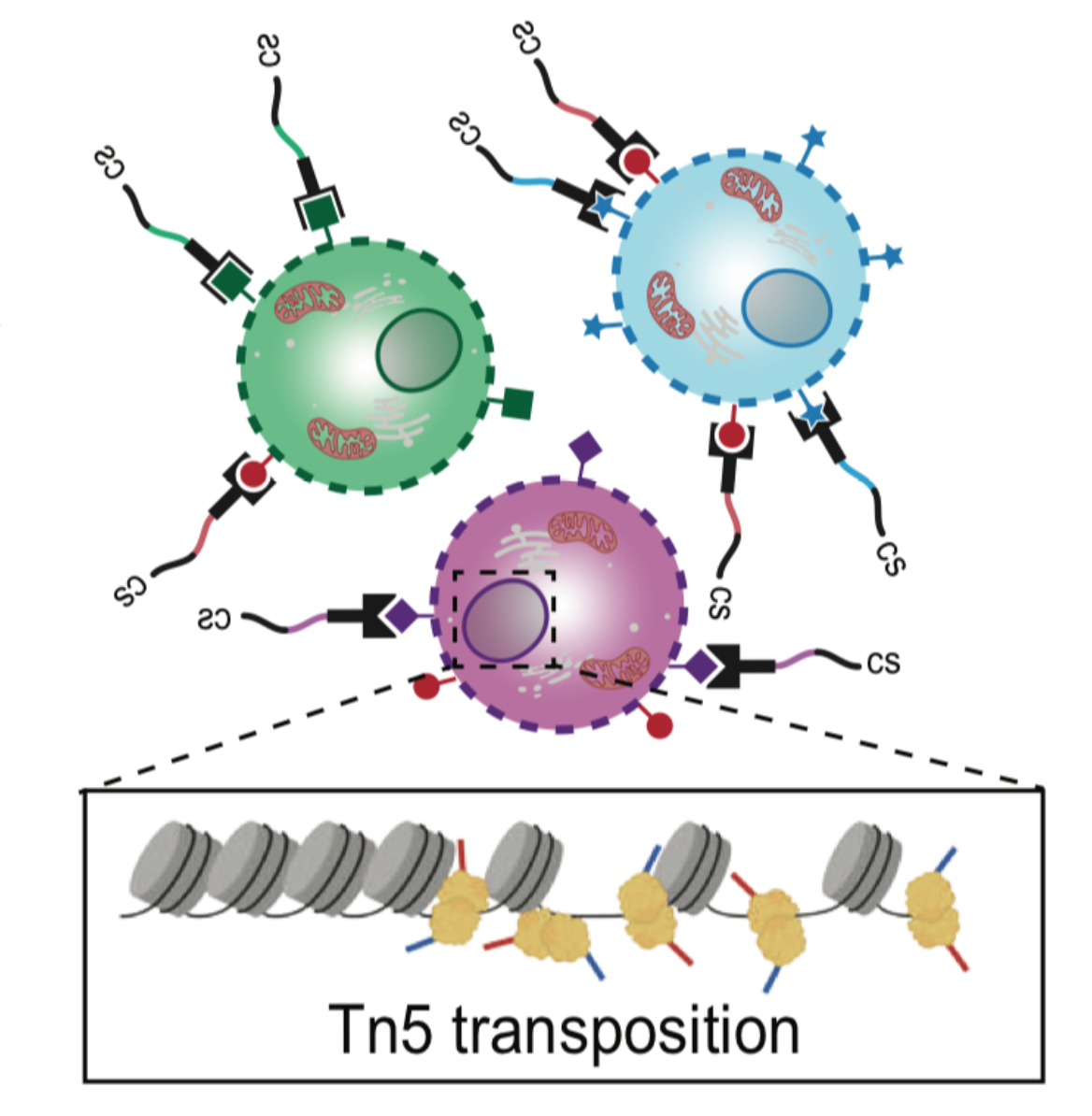
Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells
Eleni P. Mimitou, Caleb A. Lareau, Kelvin Y. Chen, Andre L. Zorzetto-Fernandes, Yuhan Hao, Yusuke Takeshima, Wendy Luo, Tse-Shun Huang, Bertrand Z. Yeung, Efthymia Papalexi, Pratiksha I. Thakore, Tatsuya Kibayashi, James Badger Wing, Mayu Hata, Rahul Satija, Kristopher L. Nazor, Shimon Sakaguchi, Leif S. Ludwig, Vijay G. Sankaran, Aviv Regev, Peter Smibert
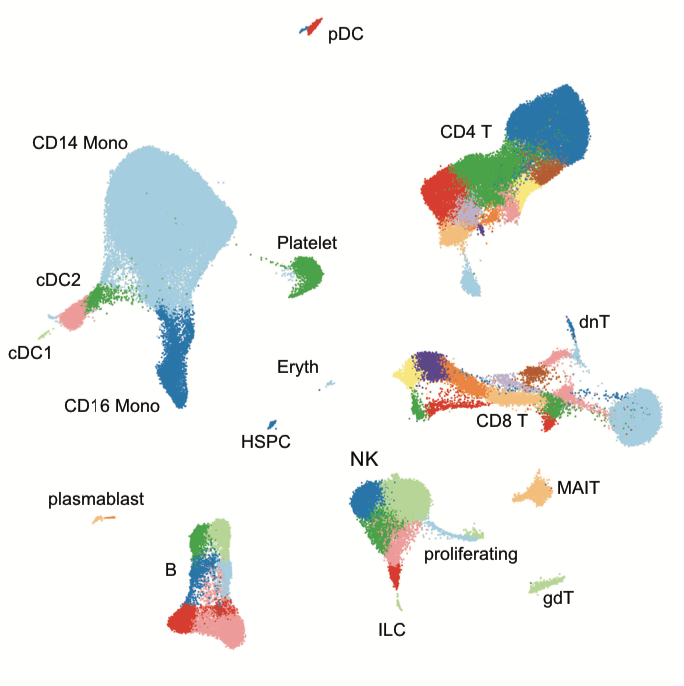
Integrated analysis of multimodal single-cell data
Yuhan Hao*, Stephanie Hao*, Erica Andersen-Nissen, William M. Mauck, Shiwei Zheng, Andrew Butler, Maddie Jane Lee, Aaron J. Wilk, Charlotte Darby, Michael Zagar, Paul Hoffman, Marlon Stoeckius, Efthymia Papalexi, Eleni P. Mimitou, Jaison Jain, Avi Srivastava, Tim Stuart, Lamar B. Fleming, Bertrand Yeung, Angela J. Rogers, Juliana M. McElrath, Catherine A. Blish, Raphael Gottardo, Peter Smibert*, Rahul Satija*

Characterizing the molecular regulation of inhibitory immune checkpoints with multi-modal single-cell screens
Efthymia Papalexi, Eleni Mimitou, Andrew W. Butler, Samantha Foster, Bernadette Bracken, William M. Mauck III, Hans-Hermann Wessels, Bertrand Z. Yeung, Peter Smibert, Rahul Satija